Data Portal for Single Cell Sequencing
Build an interactive viewer for single-cell RNA sequencing data using public datasets from the CZ CELLxGENE data portal.
Organise project
-
Create a folder for your project on your computer
-
Create a data folder outside of your project folder
Get single-cell data
-
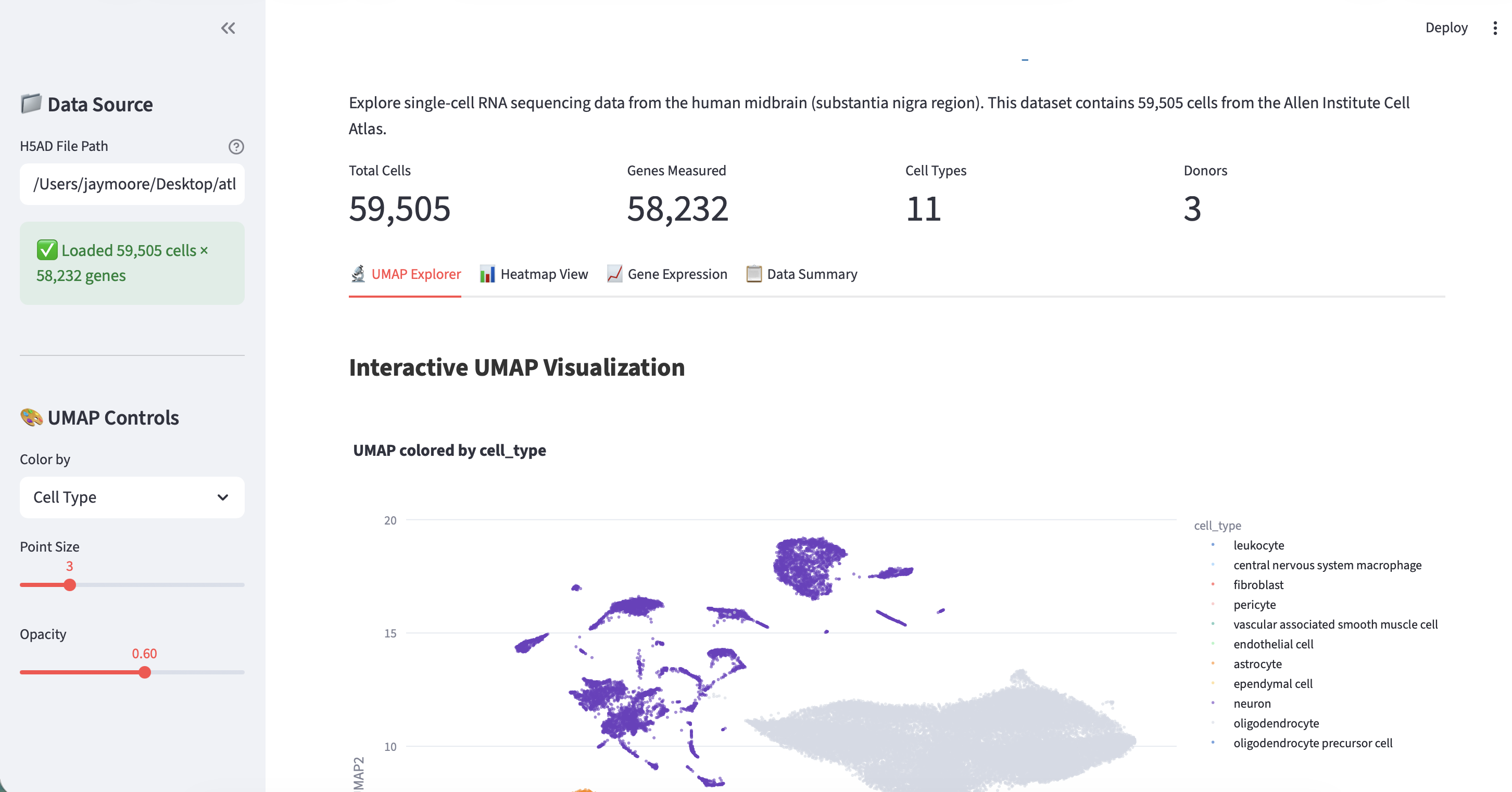
Browse the CELLxGENE collections and find a dataset. For this example we use the Allen Institute Adult Human Brain Atlas - specifically the midbrain (substantia nigra) snRNA-seq dataset.
-
Download the
.h5adfile to your laptop (it is 416.2 MB). The dataset contains 59,505 cells x 58,232 genes with 11 cell types across 3 donors. Key metadata columns includecell_type,cluster_id,supercluster_term,donor_id, and UMAP coordinates inobsm['X_UMAP'].
Get prompting!
- In
Planmode, type:
Build an interactive viewer for my single-cell RNA data. The file is at [path/to/data.h5ad].
Result
Publish to GitHub
Once your app is working, you can save it to GitHub:
Install GitHub CLI
- Download GitHub CLI
- Authenticate:
gh auth login
Upload your code
In chat mode, ask:
Push my project to a new GitHub repository called [your-repo-name]
Or create a repo first:
Create a new private GitHub repository called [your-repo-name], then push my code to it