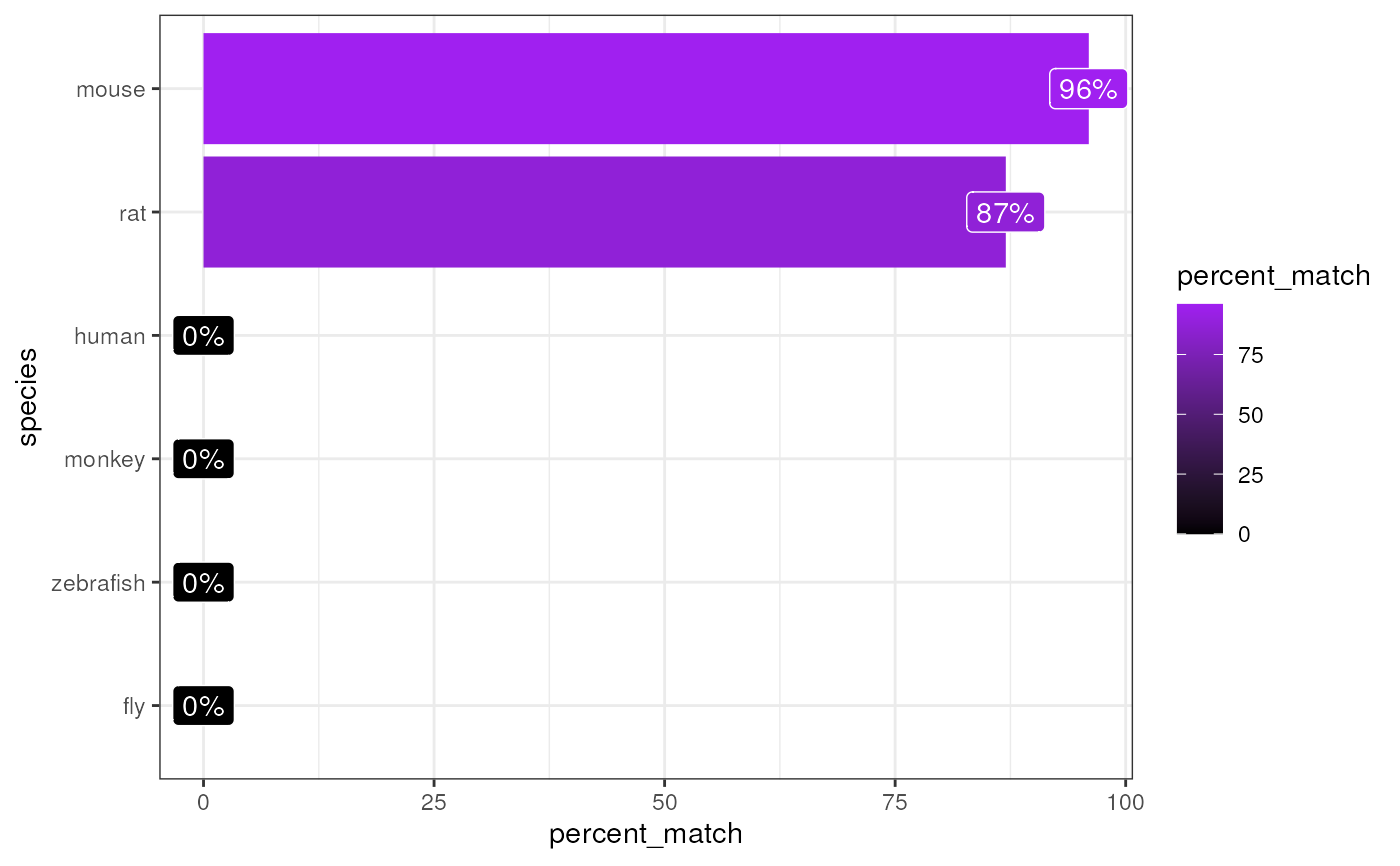
Infers which species the genes within gene_df is from.
Iteratively test the percentage of gene_df genes that match with
the genes from each test_species.
Arguments
- gene_df
Data object containing the genes (see
gene_inputfor options on how the genes can be stored within the object).
Can be one of the following formats:matrixA sparse or dense matrix.
data.frameA
data.frame,data.table. ortibble.listA
listor charactervector.
Genes, transcripts, proteins, SNPs, or genomic ranges can be provided in any format (HGNC, Ensembl, RefSeq, UniProt, etc.) and will be automatically converted to gene symbols unless specified otherwise with the
...arguments.
Note: If you setmethod="homologene", you must either supply genes in gene symbol format (e.g. "Sox2") OR setstandardise_genes=TRUE.- gene_input
Which aspect of
gene_dfto get gene names from:"rownames"From row names of data.frame/matrix.
"colnames"From column names of data.frame/matrix.
<column name>From a column in
gene_df, e.g."gene_names".
- test_species
Which species to test for matches with. If set to
NULL, will default to a list of humans and 5 common model organisms. Iftest_speciesis set to one of the following options, it will automatically pull all species from that respective package and test against each of them:- "homologene"
20+ species (default)
- "gprofiler"
700+ species
- "babelgene"
19 species
- method
R package to use for gene mapping:
"gprofiler"Slower but more species and genes.
"homologene"Faster but fewer species and genes.
"babelgene"Faster but fewer species and genes. Also gives consensus scores for each gene mapping based on a several different data sources.
- make_plot
Make a plot of the results.
- show_plot
Print the plot of the results.
- verbose
Print messages.
Value
An ordered dataframe of test_species
from best to worst matches.
Examples
data("exp_mouse")
matches <- orthogene::infer_species(gene_df = exp_mouse[1:200,])
#> Preparing gene_df.
#> sparseMatrix format detected.
#> Extracting genes from rownames.
#> 200 genes extracted.
#> Testing for gene overlap with: human
#> Retrieving all genes using: homologene.
#> Retrieving all organisms available in homologene.
#> Mapping species name: human
#> Common name mapping found for human
#> 1 organism identified from search: 9606
#> Using cached file: /github/home/.cache/R/orthogene/all_genes-9606-homologene.csv.gz
#> Returning all 19,129 genes from human.
#> Testing for gene overlap with: monkey
#> Retrieving all genes using: homologene.
#> Retrieving all organisms available in homologene.
#> Mapping species name: monkey
#> Common name mapping found for monkey
#> 1 organism identified from search: 9544
#> Using cached file: /github/home/.cache/R/orthogene/all_genes-9544-homologene.csv.gz
#> Returning all 16,843 genes from monkey.
#> Testing for gene overlap with: rat
#> Retrieving all genes using: homologene.
#> Retrieving all organisms available in homologene.
#> Mapping species name: rat
#> Common name mapping found for rat
#> 1 organism identified from search: 10116
#> Using cached file: /github/home/.cache/R/orthogene/all_genes-10116-homologene.csv.gz
#> Returning all 20,616 genes from rat.
#> Testing for gene overlap with: mouse
#> Retrieving all genes using: homologene.
#> Retrieving all organisms available in homologene.
#> Mapping species name: mouse
#> Common name mapping found for mouse
#> 1 organism identified from search: 10090
#> Using cached file: /github/home/.cache/R/orthogene/all_genes-10090-homologene.csv.gz
#> Returning all 21,207 genes from mouse.
#> Testing for gene overlap with: zebrafish
#> Retrieving all genes using: homologene.
#> Retrieving all organisms available in homologene.
#> Mapping species name: zebrafish
#> Common name mapping found for zebrafish
#> 1 organism identified from search: 7955
#> Using cached file: /github/home/.cache/R/orthogene/all_genes-7955-homologene.csv.gz
#> Returning all 20,897 genes from zebrafish.
#> Testing for gene overlap with: fly
#> Retrieving all genes using: homologene.
#> Retrieving all organisms available in homologene.
#> Mapping species name: fly
#> Common name mapping found for fly
#> 1 organism identified from search: 7227
#> Using cached file: /github/home/.cache/R/orthogene/all_genes-7227-homologene.csv.gz
#> Returning all 8,438 genes from fly.
#> Top match:
#> - species: mouse
#> - percent_match: 96%