Import a phylogenetic tree and then conduct
a series of optional standardisation steps.
Optionally, if output_format is not NULL, species names from
both the tree and the species argument will first be standardised
using map_species.
Arguments
- tree_source
Can be one of the following:
- "timetree2022":
Import and prune the TimeTree >147k species phylogenetic tree. Can also simply type "timetree".
- "timetree2015":
Import and prune the TimeTree >50k species phylogenetic tree.
- "OmaDB":
Construct a tree from OMA (Orthologous Matrix browser) via the getTaxonomy function. NOTE: Does not contain branch lengths, and therefore may have limited utility.
- "UCSC":
Import and prune the UCSC 100-way alignment phylogenetic tree (hg38 version).
- "<path>":
Read a tree from a newick text file from a local or remote URL using read.tree.
- "timetree2022":
- species
Species names to subset the tree by (after
standardise_speciesstep).- output_format
Which column to return.
- run_map_species
Whether to first standardise species names with map_species.
- method
R package to use for gene mapping:
"gprofiler"Slower but more species and genes.
"homologene"Faster but fewer species and genes.
"babelgene"Faster but fewer species and genes. Also gives consensus scores for each gene mapping based on a several different data sources.
- force_ultrametric
Whether to force the tree to be ultrametric (i.e. make all tips the same date) using force.ultrametric.
- age_max
Rescale the edges of the tree into units of millions of years (MY) instead than evolutionary rates (e.g. dN/dS ratios). Only used if
age_max, the max number , is numeric. Times are computed using makeChronosCalib and chronos.- show_plot
Show a basic plot of the resulting tree.
- save_dir
Directory to cache full tree in. Set to
NULLto avoid using cache.- verbose
Print messages.
- ...
Additional arguments passed to makeChronosCalib.
Value
A filtered tree of class "phylo" (with standardised species names).
Examples
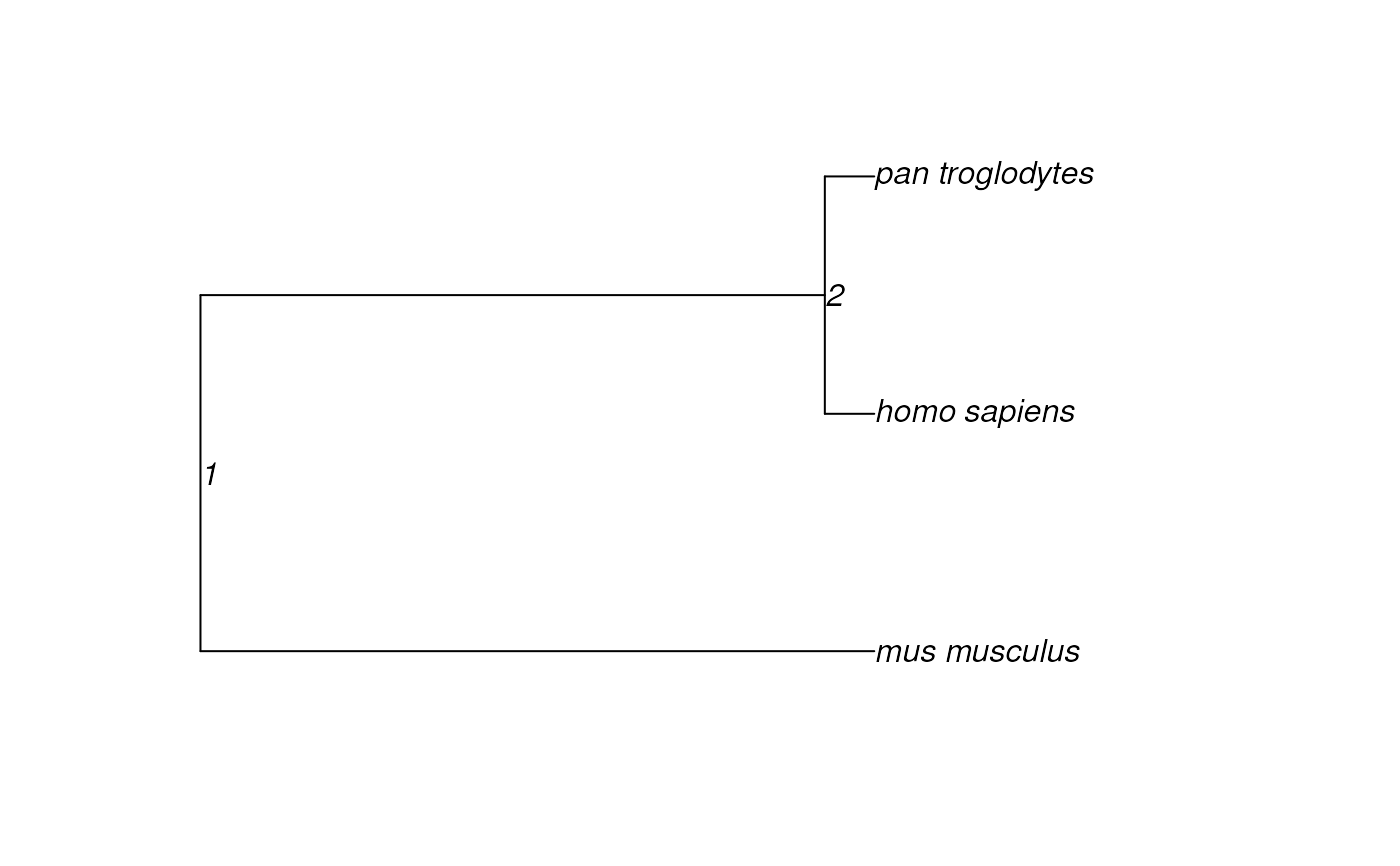
if(require("ape")){
species <- c("human","chimp","mouse")
tr <- orthogene::prepare_tree(species = species)
}
#> Importing tree from: TimeTree2022
#> Importing cached tree.
#> Standardising tip labels.
#> Mapping 3 species from `species`.
#> Mapping 3 species from tree.
#> --
#> 0/3 (0%) tips dropped from tree due to inability to standardise names with `map_species`.
#> --
#> 0/3 (0%) tips dropped from tree according to overlap with selected `species`.