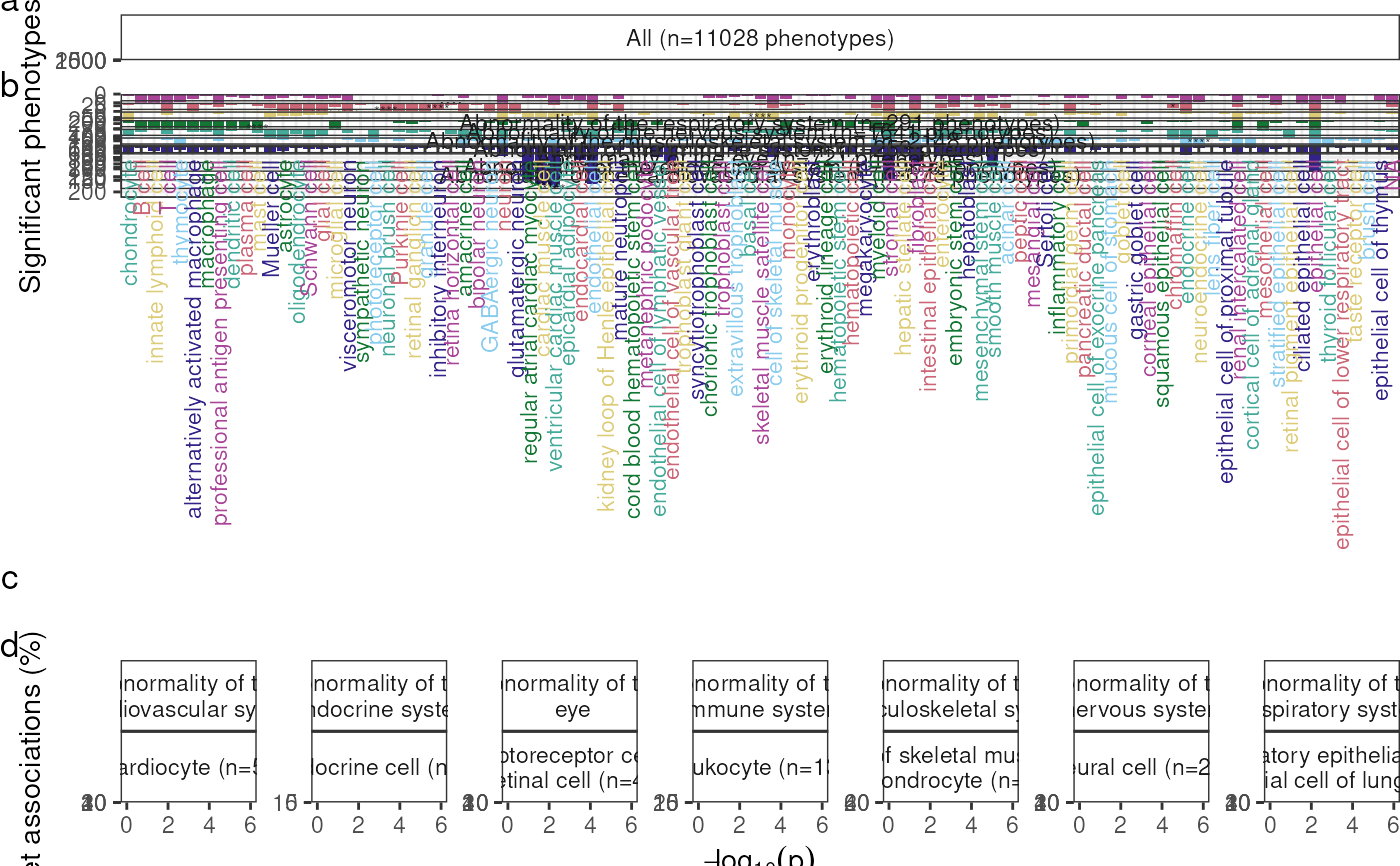
Create a plot summarising MSTExplorer results as a bar chart with multiple facets showing selected branches from the Human Phenotype Ontology (HPO). Also shows a dendrogram of celltype-celltype relationships using the Cell Ontology (CL).
plot_bar_dendro(
results = load_example_results(),
celltype_col = "cl_name",
target_branches = get_target_branches(),
keep_ancestors = names(target_branches),
hpo = HPOExplorer::get_hpo(),
cl = get_cl(),
facets = "ancestor_name",
add_test_target_celltypes = TRUE,
add_prop_test = FALSE,
preferred_palettes = "tol",
legend.position = "none",
heights = c(0.3, 1, 0.15, 0.3),
expand_dendro_x = rep(0.01, 2),
q_threshold = 0.05,
show_plot = TRUE,
save_path = NULL,
height = 16,
width = 13
)Arguments
- results
The cell type-phenotype enrichment results generated by gen_results and merged together with merge_results
- celltype_col
Name of the cell type column in the
results.- target_branches
A named list of HPO branches each matched with CL cell type branches that correspond to on-target cell types across the two ontologies.
- keep_ancestors
Only HPO terms that have these ancestors will be kept.
- hpo
Human Phenotype Ontology object, loaded from get_ontology.
- cl
Cell Ontology (CL) object from
KGExplorer::get_ontology("cl").- facets
A set of variables or expressions quoted by
vars()and defining faceting groups on the rows or columns dimension. The variables can be named (the names are passed tolabeller).For compatibility with the classic interface, can also be a formula or character vector. Use either a one sided formula,
~a + b, or a character vector,c("a", "b").- add_test_target_celltypes
Using the significant phenotype-cell type association
results, run proportional enrichment tests to determine whether each cell type is overrepresented in a given HPO branch relative to all other HPO branches. Overrepresented cell types will be denoted by "*" above its bar.- add_prop_test
Add proportional enrichment results using
run_prop_tests.- preferred_palettes
Preferred palettes to use for each column.
- legend.position
the default position of legends ("none", "left", "right", "bottom", "top", "inside")
- heights
Passed to wrap_plots.
- expand_dendro_x
Passed to scale_x_discrete in the cell type dendrogram.
- q_threshold
The q value threshold to subset the
resultsby.- show_plot
Print the plot to the console.
- save_path
Save the plot to a file. Set to
NULLto not save the plot.- height
Height of the saved plot.
- width
Width of the saved plot.
Value
A bar chart with dendrogram of EWCE results in each cell type.
Examples
results <- load_example_results()
out <- plot_bar_dendro(results = results)
#> Loading required namespace: ggdendro
#> Adding HPO names.
#> Translating ontology terms to names.
#> Adding level-2 ancestor to each HPO ID.
#> Adding ancestor metadata.
#> Ancestor metadata already present. Use force_new=TRUE to overwrite.
#> 2,206,994 associations remain after filtering.
#> Mapping cell types to cell ontology terms.
#> Adding stage information.
#> Filtered 'ancestor_name' : 999,488 / 2,206,994 rows dropped.
#> Translating ontology terms to ids.
#> Translating ontology terms to ids.
#> Using cached ontology file (1/1):
#> /github/home/.cache/R/KGExplorer/ontologies/github/cl_v2023-09-21.rds
#> Keeping descendants of 2 term(s).
#> 2,711 terms remain after filtering.
#> Translating ontology terms to ids.
#> Translating ontology terms to ids.
#> Translating ontology terms to ids.
#> Translating ontology terms to ids.
#> Translating ontology terms to ids.
#> Translating ontology terms to ids.
#> Translating ontology terms to ids.
#> Using cached ontology file (1/1):
#> /github/home/.cache/R/KGExplorer/ontologies/github/cl_v2023-09-21.rds
#> Keeping descendants of 2 term(s).
#> 2,711 terms remain after filtering.
#> Translating ontology terms to ids.
#> Translating ontology terms to ids.
#> Converted ontology to: igraph
#> Converted ontology to: igraph_dist
#> Translating ontology terms to names.
#> Adding logFC column.
#> Using palette: tol
#> Cell type columns already present. Skipping mapping.
#> Ancestor columns already present. Skipping.
#> 2,206,994 associations remain after filtering.
#> Cell type columns already present. Skipping mapping.
#> Running tests: across_branches_per_celltype
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.
#> Scale for y is already present.
#> Adding another scale for y, which will replace the existing scale.
#> Ancestor columns already present. Skipping.
#> 2,206,994 associations remain after filtering.
#> Cell type columns already present. Skipping mapping.
#> Translating ontology terms to ids.
#> Translating ontology terms to ids.
#> Using cached ontology file (1/1):
#> /github/home/.cache/R/KGExplorer/ontologies/github/cl_v2023-09-21.rds
#> Keeping descendants of 2 term(s).
#> 2,711 terms remain after filtering.
#> Translating ontology terms to ids.
#> Translating ontology terms to ids.
#> Translating ontology terms to ids.
#> Translating ontology terms to ids.
#> Translating ontology terms to ids.
#> Translating ontology terms to ids.
#> Proportional enrichment summary stats:
#> - pct_min: 17.22
#> - pct_max: 64.86
#> - pct_max_mean: 39.9
#> - pct_max_sd: 17.66
#> - enrichment_mean: 4.75
#> Warning: Arguments in `...` must be used.
#> ✖ Problematic argument:
#> • na.rm = TRUE
#> ℹ Did you misspell an argument name?